DMcloud is a macromolecular modeling tool for local structure fitting using point cloud matching with a diffusion model for cryo-EM maps at < 10Å resolution
It is especially useful when fitting models, such as those from AlphaFold, that may be globally incorrect but contain accurate local structures.
If encounter problems, please contact Daisuke Kihara (dkihara@purdue.edu) or Xiao Wang (xiaowang20140001@gmail.com) or Genki Terashi (gterashi@purdue.edu)
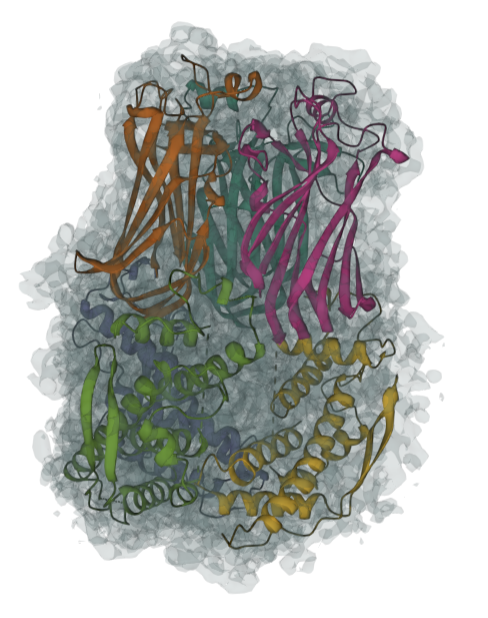
Example EMD-1461
Input Map file: emd_1461.mrc
Input AF2 template file: emd_2513_af2.pdb
Contour level: 0.05
Result Example:Result Example with full-atom model building
Please simply click "Upload" when you filled all input fields.
Density Histogram (|ρ|)
0.00
Contour: 0.00
0.00
Please do not input 0, you must provide a contour to remove the outside very noisy regions
Please refer to the instructions for How to Preparing the PDB file. If your target is a hetero-oligomer and you have multiple PDB files, please upload all of them
Are the PDB files from AlphaFold? If you have multiple protein structures, and some are from AlphaFold while others are not, please select "No"
0/500
This note will help you distinguish jobs later.