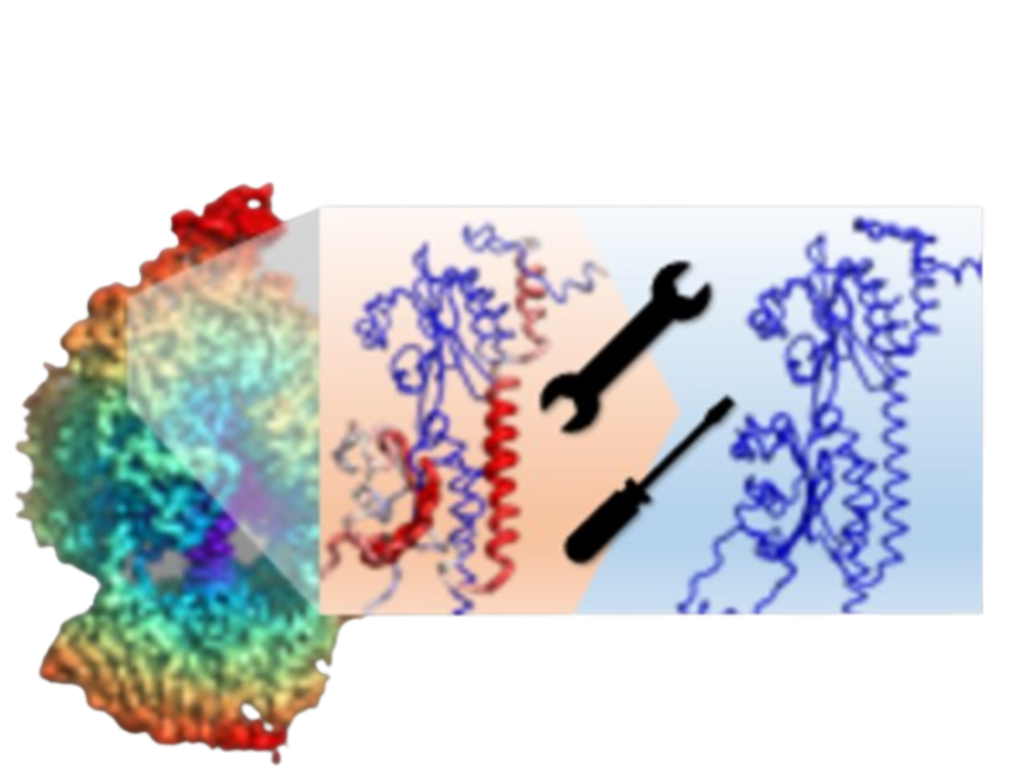
DAQ is a computational tool employing deep learning to estimate residue-wise local quality for
protein models derived from cryo-EM maps (target resolution: 0-5 A).
It generates a file in .pdb format documenting the scored structure, with scores stored in the
B-factor column
Our result webpage comprises three tabs: Results Visualization, Output Logs, and Job Configuration.
The 'Result Visualization' panel showcases the protein structure scored by DAQ-Score, with the
DAQ(AA) score stored in the b-factor column.
The 3D model is color-coded based on the DAQ(AA) score, ranging from red (-1.0) to blue (1.0).
Blue represents a good score, while red signifies a lower score from DAQ.
On the right-hand side, you'll find the 'Download Outputs' button to acquire the modeled structure
in .pdb format.
You can utilize 'spectrum b, red_white_blue, all, -1,1' in PyMol to visualize the score.
Additionally, you can visualize the map online by clicking the "Show map" button.
Once loaded, the default contour level matches your input; however, you can make adjustments by
clicking the "..." button beside "isosurface."
Within the "Type: Isosurface" option, you can modify the iso-surface value and opacity by scrolling
through the bar for precise adjustments.
This feature allows you to assess the alignment between the modeled structure and the map.
Output Logs:
The 'Output Logs' panel compiles all outputs generated by the scripts.
If you're interested in monitoring the job's progress during execution, this section provides a
comprehensive overview.
Job Configuration:
In the 'Job Configuration' panel, you'll find the input parameters used for this specific job.
These records serve to maintain a log of your submitted input for reference.
Problem Debugging:
For any troubleshooting needs: Should you encounter any issues, please don't hesitate to contact
us via
email to report the problems.
When sending an email, kindly use the subject line format 'DAQ-score problem: [jobid]', where
[jobid]
corresponds to the job displayed in the title.
This specific identification helps us efficiently locate and debug jobs in the backend, ensuring
a prompt response to your concerns.
Contact:
dkihara@purdue.edu, gterashi@purdue.edu, xiaowang20140001@gmail.com.
DAQ Score