Emap2sec Tutorial
Overview
Emap2sec is a computational tool that can identify protein secondary structures in cryo-Electron Microscopy (EM) maps up to 5-10 Å.
Overall Pipeline
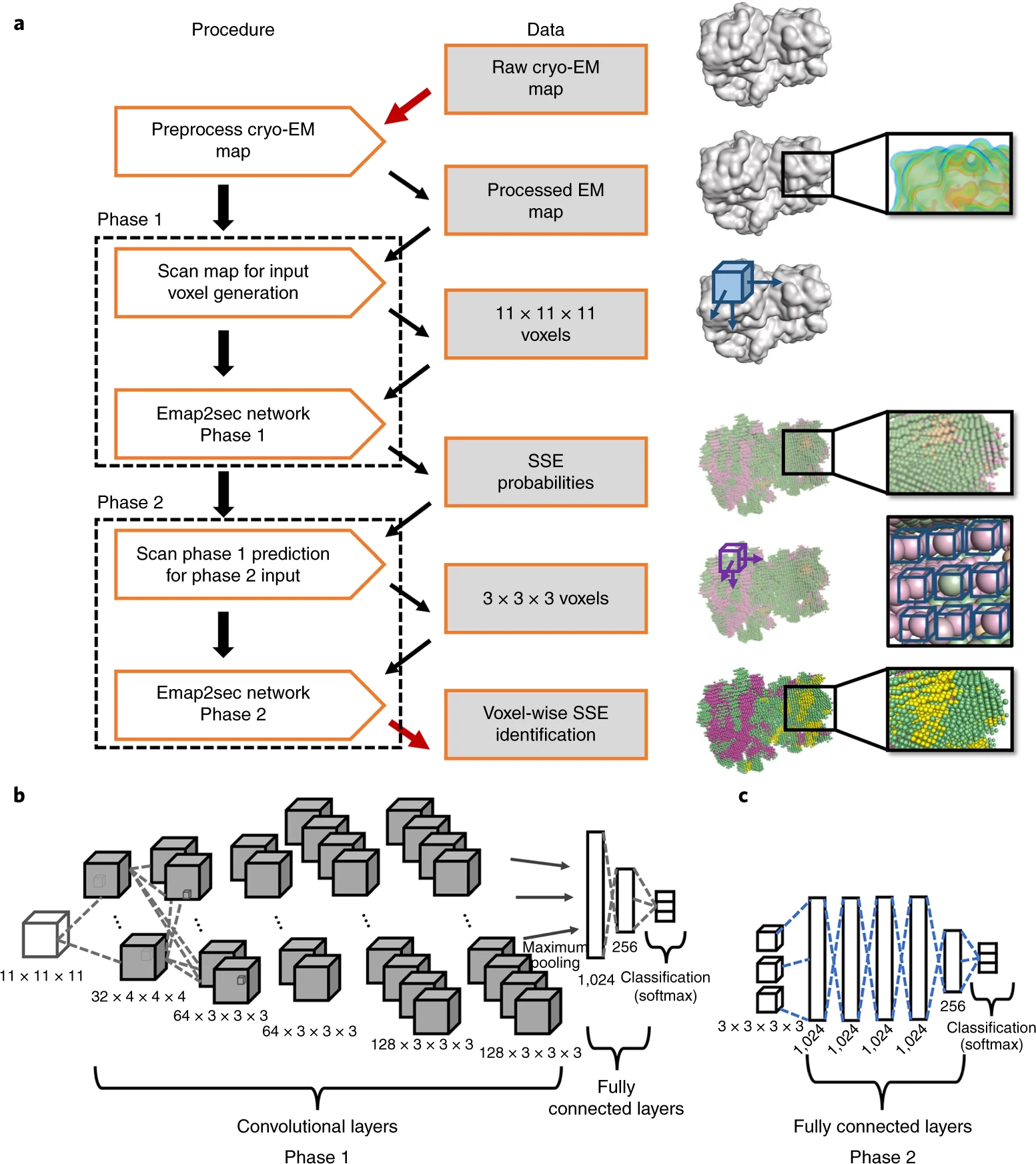
- The algorithm takes the EM map and relevant details such as the map's author recommended contour level and pixels/Angstrom values as input and generates an intermediate readable text file (trimmap) which contains the normalized electron density values of voxels.
- A program named dataset uses this trimmap to generate an input file, which contains rows of density values that we get by scanning the input map in all the 3 directions using a 11*11*11 cube. Emap2sec makes secondary structure assignment to each of these rows.
- CNN for local structure detection (Phase 1): The test dataset generated in the previous step is fed to the first phase of Emap2sec that contains a convolutional neural network (CNN). This step gives out probability values of helix, sheet, and other for each cube.
- The figure (B) shows the architecture of the phase 1 deep neural networks, consisting of five CNN layers followed by one maximum-pooling layer. The first CNN has 32 filters that are 43Å3 in size, the second and third CNNs have 64 filters that are 33Å3 in size, and the fourth and fifth CNNs have 128 filters of 33Å3 in size. The last layers of the network are two fully connected layers, which have 1,024 and 256 nodes each. The fully connected layers are connected to the output layer, which uses the softmax function to compute the probabilities for the three secondary structure classes.
- Prediction smoothing (Phase 2): The output from the first phase is a set of predicted probabilities for each of helix, sheet, and other structures. In this second phase, these probabilities are used to smooth out any infeasible predictions given in the first phase.
- The figure (C) shows the phase 2 network, consisting of five fully connected layers followed by an output layer.
- Finally, the top-ranked model by DAQ(AA) score is selected as the final model.
Input Files
3D cryo-EM map in .mrc format
Output Files
Modeled Protein Secondary structure in .pdb format
Job Submission
- Prepare Input Map
Collect 3D cryo-EM map from microscope in .mrc and .map format.
Example map
You can also find many maps in EMDataResource as testing examples.
- Decide the contour level
Please make sure your contour level is lower than your focused region. This is absolute density threshold, not standard deviation.
Please input valid contour level since it is used for normalization in Emap2sec. 0 is not suggested and may ruin the visualization. - Submit your job
Once you collected the input files, please submit your job here. For each input field, please input the files/info collected before.
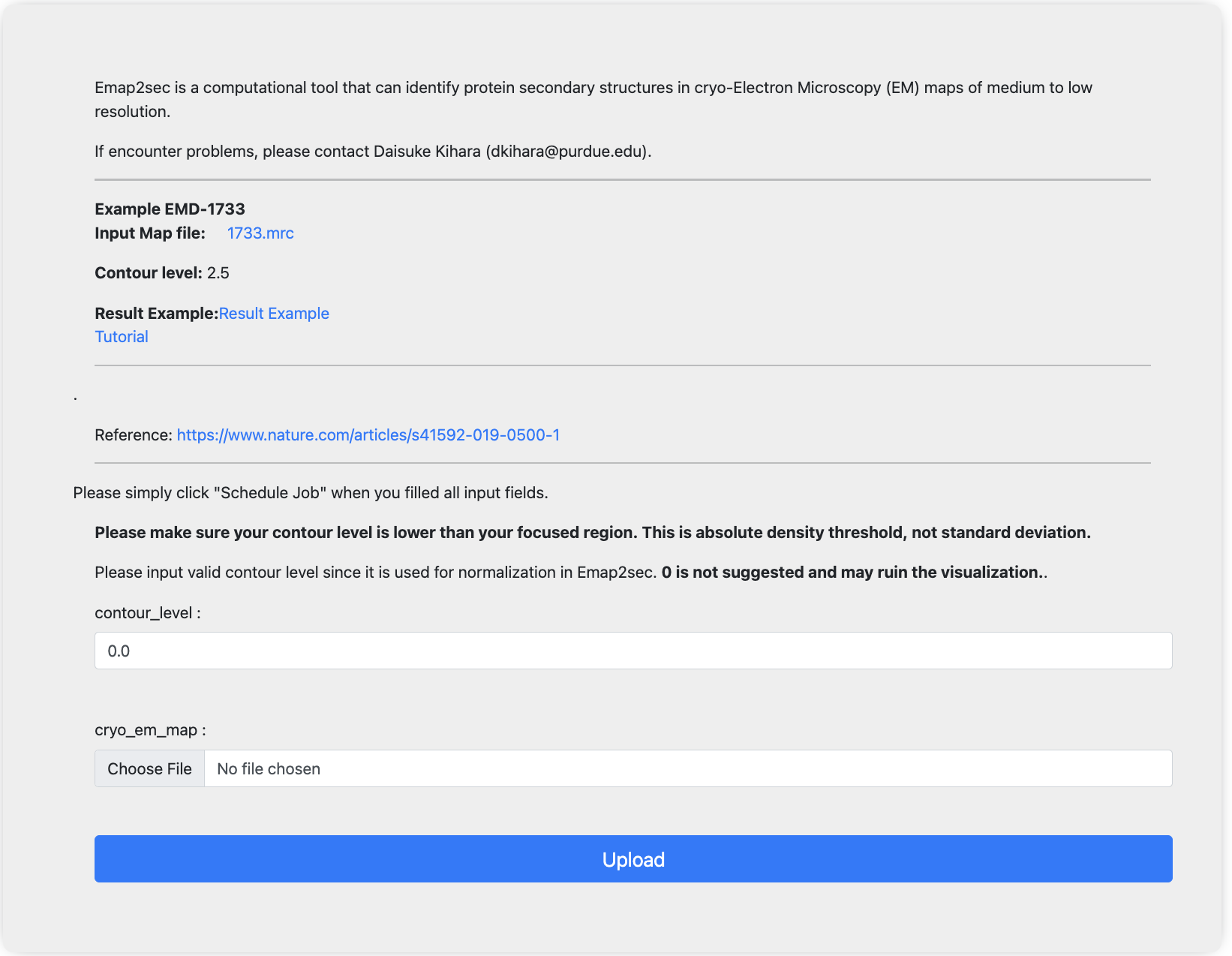
Once you finished input, simply click the upload button to submit jobs. After submission, you will be redirected to the “view job“ page. If you are not registered, please bookmark the link. Once the job is done, you can view jobs from this link. If you are registered, you will receive email notifications once job is done and you can also check job status from my jobs list under job manager.
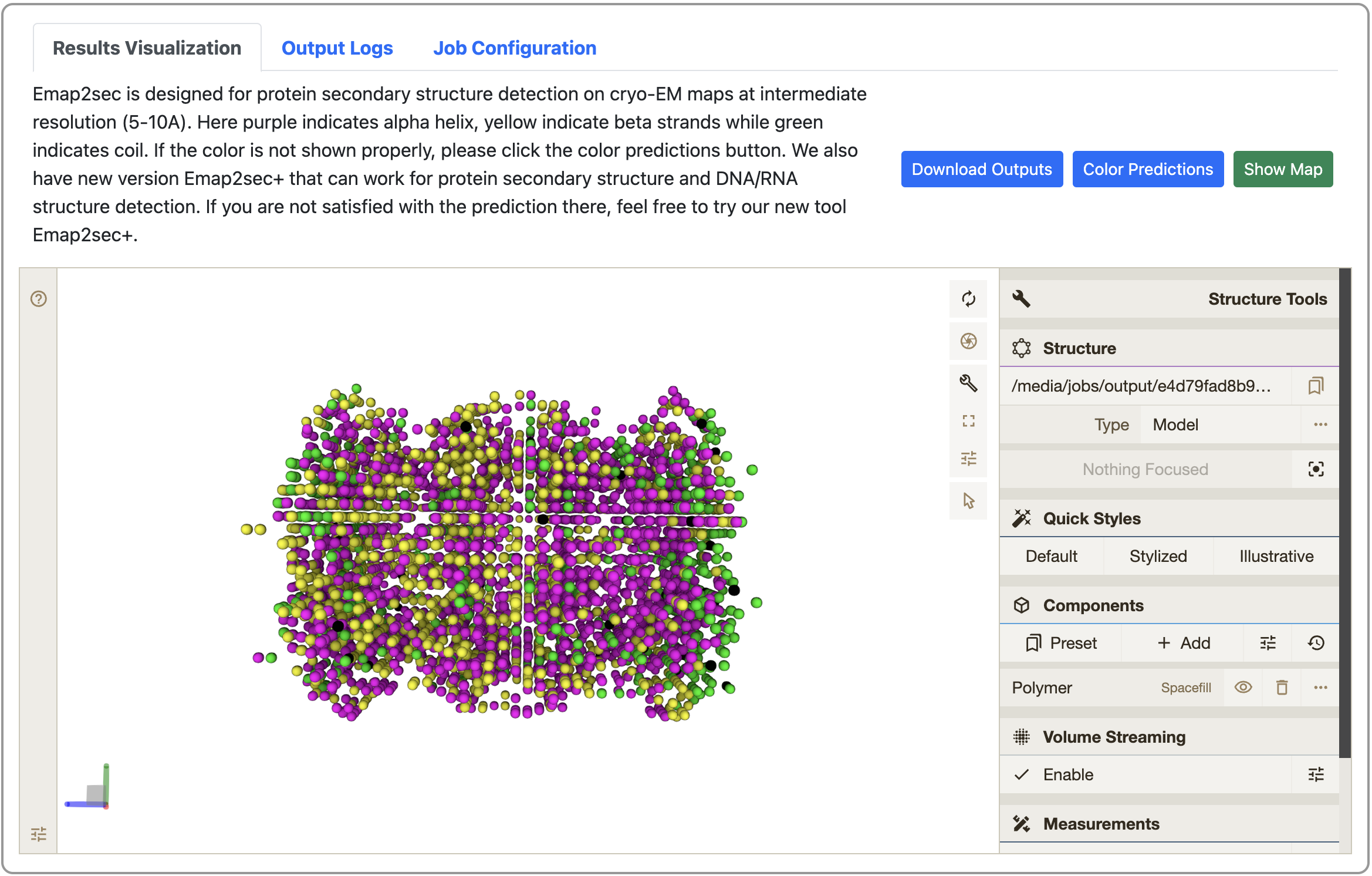
- View your job results
Once job is done, you can check the modeled structure from the link bookmarked before. Here you can also download the modeled structure in .pdb format by clicking the “Download Outputs” button. You can also visualize the 3D cryo-EM map online to check its consistency with the modeled structure. For more detailed instructions, please see the “Instructions” in the same page.
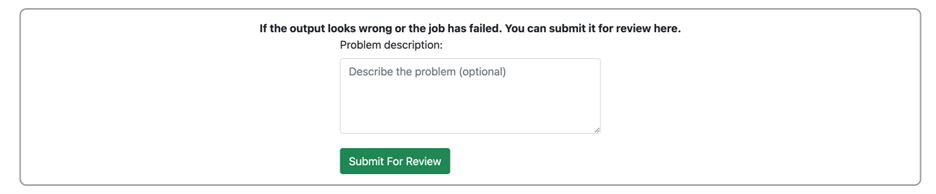
- Submit for backend review (optional)
If you noticed any strange outputs or job failure on your side, please submit a backend review by using the field in the bottom of the “View Job” page. We will get back to you as soon as possible.
Availability (Other)
- GitHub
This github contains a modified ColabFold notebook and our tools.
- Google
Colab
Step-by-step instructions are available.
Reference
Maddhuri Venkata Subramaniya, S. R., Terashi, G., & Kihara, D. (2019). Protein secondary structure detection in intermediate-resolution cryo-EM maps using deep learning. Nature Methods, 16(9), 911-917. https://www.nature.com/articles/s41592-019-0500-1